Implen Journal Club | December Issue |
|
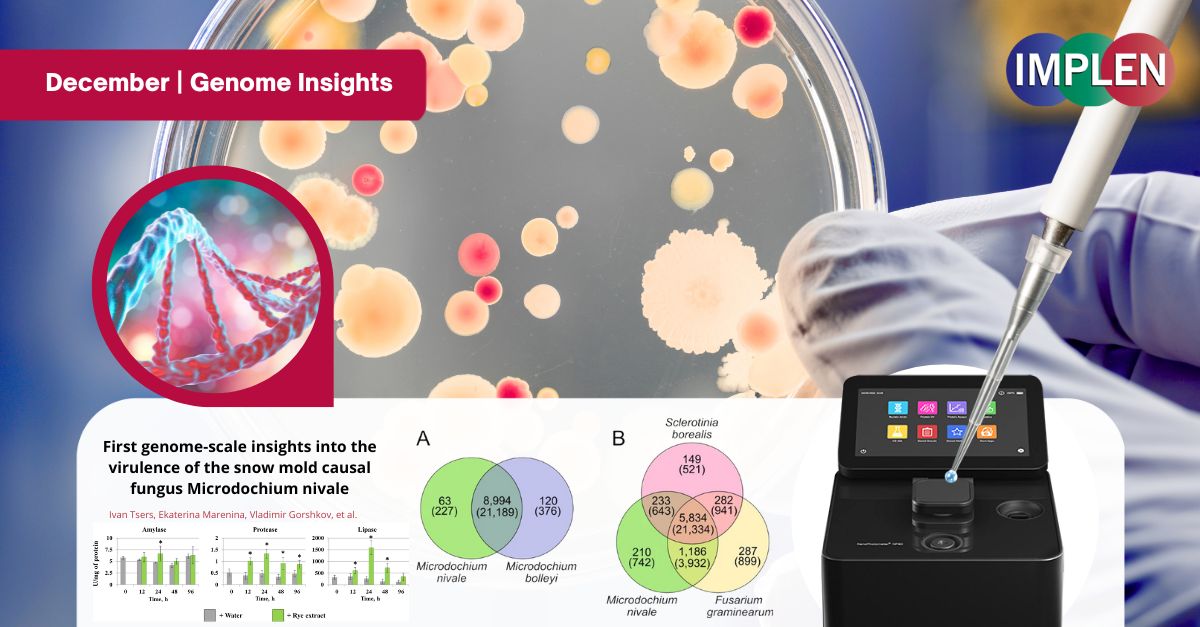
Genome insights into the virulence of the snow mold causal fungusWith snowfall on the way for many, the first issue of the Implen NanoPhotometer® Journal Club is featuring research published by Tsers et al. on Pink snow mold (Microdochium nivale), which poses a significant threat to winter crops including rye. This study investigated the fungus's genome, revealing its ability to produce harmful mycotoxins, a potential factor in its virulence. It was found that pink snow mold adapts to its environment by altering its metabolism in response to plant chemicals, enhancing its ability to infect plants. Pink snow mold thrives in snowy conditions, making it a major concern in areas with prolonged snowfall. By sequencing its genome, new insights were gained into the mechanisms that allow pink snow mold to survive and damage crops under snow cover. This study demonstrated that the fungus can trigger enzymes that break down plant material, enabling it to spread and harm crops. Additionally, the genetic potential for producing mycotoxins, previously unknown, was uncovered in this research. Understanding the genetic makeup of pink snow mold and how it interacts with its host plants will be important for managing snow mold outbreaks and protecting crops during the winter months. This research has the potential to inform better strategies for controlling the spread of this destructive pathogen. The Implen NanoPhotometer® was used in this study to assess the quality and quantity of DNA. |
|
Nutcracker Science: Unlocking the Wonders of Almond-Derived NanovesiclesAs the festive month of December unfolds, bringing almonds into the spotlight in seasonal treats and traditions, this week’s Impen NanoPhotometer Journal Club explores the fascinating world of almond-derived nanovesicles (ADNVs) and their potential health benefits. A study recently published by Santangelo et al. investigated into the extraction of these tiny structures from raw, roasted, and blanched almonds, revealing their small size (100-200 nanometers), round shape, and stability in liquids due to their slight negative charge. ADNVs contained proteins including TET8, which suggested they might help in communication between species and act as carriers for beneficial compounds. Even after heat heating by roasting or blanching—common preparations for almonds—these nanovesicles remained intact, showing resilience that made them ideal for drug delivery systems. Their natural, safe, and body-compatible properties opened the door for exciting health applications. The study also highlighted the potential for using almond byproducts to collect these vesicles on a larger scale, turning food processing waste into valuable resources. This research showed almonds as not only a healthy food and festive favorite but also a key player in advancing nanotechnology and medicine. The Implen NanoPhotometer® NP80 was used in the research to evaluate the purity of the extracellular vesicle samples (Ev's). |
|
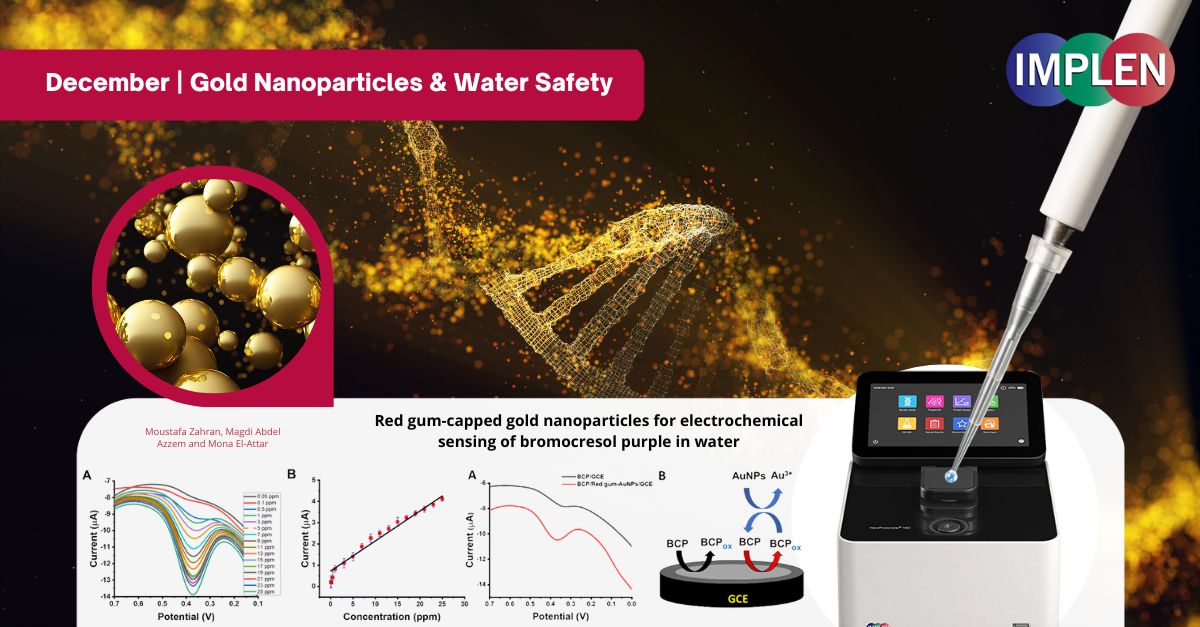
Revolutionizing Water Safety: How Gold Nanoparticles Are Transforming Environmental ResearchNext issue is is celebrating the "12 Days of Christmas" by showcasing the innovative work of Zahran et. al. on gold nanoparticles. This study introduced a novel approach to detecting bromocresol purple (BCP), a dye harmful to aquatic ecosystems, using gold nanoparticles (AuNPs) capped with red gum from Eucalyptus tereticornis. BCP alters water properties, posing risks to aquatic life, underscoring the need for sensitive detection methods. This work synthesized the nanoparticles by reducing gold(III) with red gum, yielding stable, spherical particles (~25 nm) confirmed through advanced characterization techniques. These AuNPs were then applied to modify a glassy carbon electrode (GCE), enhancing its surface area and catalytic properties. The modified sensor demonstrated exceptional sensitivity, using square-wave voltammetry (SWV) to detect BCP concentrations as low as 0.015 ppm with 99% recovery in real water samples. Critical parameters such as pH and scan time were optimized for maximum performance. Notably, the sensor exhibited strong resistance to interference from other substances, ensuring reliability in complex water environments. This fast, cost-effective, and highly sensitive method provides a valuable tool for monitoring and mitigating the impact of harmful dyes, offering a practical solution to protect aquatic ecosystems. The NanoPhotometer® N60 was used in this work to optically identify and characterize the red gum-AuNPs using the wavelength range of 200–900 nm to monitor the characteristic peak of AuNPs. |
|
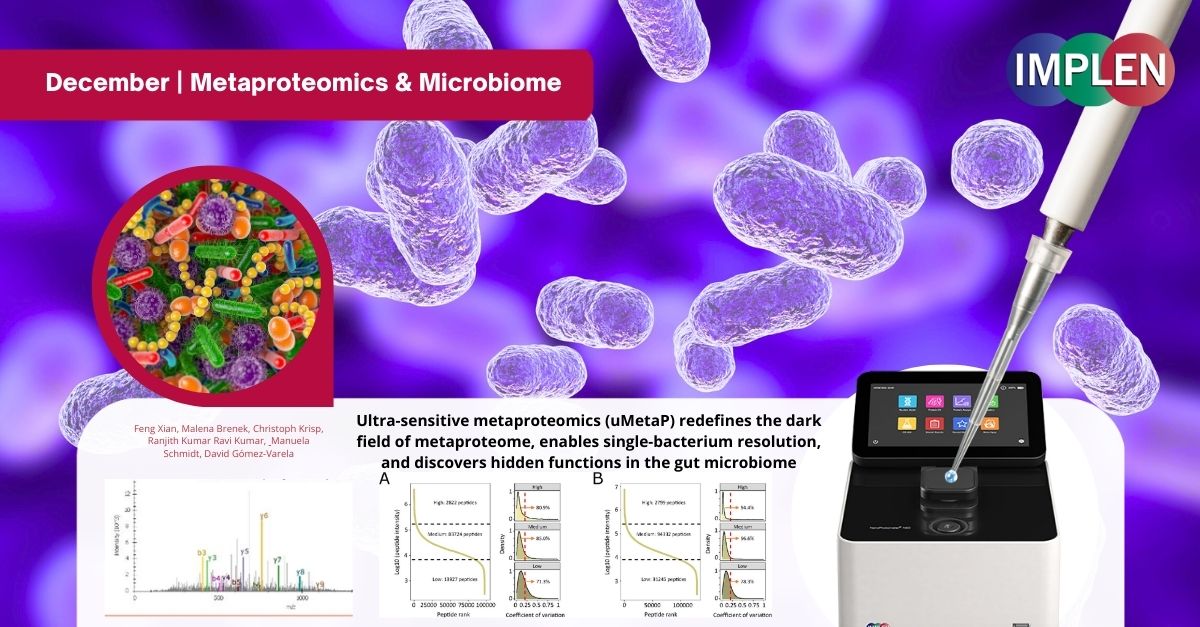
Unwrapping the Microbial Mystery: A Quantum Leap in Metaproteomics with uMetaPOur last December issue continues the celebration of the "12 Days of Christmas" with the work of Xian et. al. making a quantum leap in metaproteomics with the Ultra-sensitive metaproteomics (uMetaP) platform. Designed for ultra-sensitive detection and single-bacterium resolution, uMetaP unveils the hidden world of the microbiome with unprecedented precision and speed. Metaproteomics studies the proteins of microbial communities, crucial for understanding how microbes interact with hosts and ecosystems. Traditional methods often miss low-abundance species and functional insights, leaving much of the microbiome unexplored. uMetaP, a revolutionary tool combining cutting-edge techniques including DIA-PASEF and Stable Isotope Labeling in Cell Culture (SILAC) enabled detection of microbial proteins at a sensitivity up to 5,000 times greater than previous methods, offering detailed taxonomic and functional profiling. In just 30 minutes, uMetaP identified 47,925 protein groups, 220 microbial species, and 223 KEGG pathways with remarkable accuracy and reproducibility. It revealed previously undetectable proteins, including small proteins, proteins of unknown function, and potential new antibiotics. By detecting bacterial species representing as little as 0.001% of total biomass, uMetaP paves the way for early disease diagnosis and deeper insights into microbial diversity. This revolutionary platform sets a new standard in microbiome research, empowering scientists to explore microbial ecosystems with unparalleled depth and precision—truly a gift for science this holiday season. The NanoPhotometer® N60 was used to measure the peptide concentration. |
©2024 Implen. All rights reserved.